I’m happy to announce that naniar version 1.1.0 “Prince Caspian” is released. It’s been about a year since the last releaase, so I’m happy to include some new features. Let’s explore some of the new changes in this release
Imputation functions
I’ve always been a hesitant to include imputation helper functions like impute_mean(), and friends in {naniar}, as they aren’t a great tool to use for imputation. I also really believe the {simputation} R package does a great job at doing much imputation. However, sometimes you do actually want to demonstrate how bad imputing the mean is. Or sometimes you do need to impute the mean. And I’d rather make it easier for people to do this.
This release included some new functions: impute_fixed(), impute_zero(), impute_factor(), and impute_mode(). Notably, these do not implement “scoped variants” which were previously implemented - for example, impute_fixed_if() etc. This is in favour of using the new across() workflow within {dplyr}, which is a bit easier to maintain. This resolves issues #261 and #213.
To demonstrate these imputation functions, let’s create a vector of missing values:
library(naniar)
vec_num <- rnorm(10)
vec_int <- rpois(10, 5)
vec_fct <- factor(LETTERS[1:10])
vec_num <- set_prop_miss(vec_num, 0.4)
vec_int <- set_prop_miss(vec_int, 0.4)
vec_fct <- set_prop_miss(vec_fct, 0.4)
vec_num
#> [1] NA 0.9232763 NA 0.5265162 NA NA
#> [7] 1.4260531 1.3793303 -0.7855565 -0.5928170
vec_int
#> [1] NA 5 6 5 5 NA NA 6 NA 6
vec_fct
#> [1] <NA> B <NA> D <NA> F G <NA> I J
#> Levels: A B C D E F G H I J
We can use impute_fixed() to impute fixed values into the numeric of these:
impute_fixed(vec_num, -999)
#> [1] -999.0000000 0.9232763 -999.0000000 0.5265162 -999.0000000
#> [6] -999.0000000 1.4260531 1.3793303 -0.7855565 -0.5928170
impute_fixed(vec_int, -999)
#> [1] -999 5 6 5 5 -999 -999 6 -999 6
And impute_zero is just a special case of impute_fixed, where the fixed value is 0:
impute_zero(vec_num)
#> [1] 0.0000000 0.9232763 0.0000000 0.5265162 0.0000000 0.0000000
#> [7] 1.4260531 1.3793303 -0.7855565 -0.5928170
impute_zero(vec_int)
#> [1] 0 5 6 5 5 0 0 6 0 6
Similar to impute_mean(), impute_mode() imputes the mode:
impute_mode(vec_num)
#> [1] 1.0704569 0.9232763 1.0704569 0.5265162 1.0704569 1.0704569
#> [7] 1.4260531 1.3793303 -0.7855565 -0.5928170
impute_mode(vec_int)
#> [1] 6 5 6 5 5 6 6 6 6 6
You can’t however use impute_fixed() or impute_zero() on factors, and this doesn’t work for factors, even if it’s a new character.
impute_fixed(vec_fct, -999)
#> Warning in `[<-.factor`(`*tmp*`, is.na(x), value = -999): invalid factor level, NA generated
#> [1] <NA> B <NA> D <NA> F G <NA> I J
#> Levels: A B C D E F G H I J
impute_zero(vec_fct)
#> Warning in `[<-.factor`(`*tmp*`, is.na(x), value = 0): invalid factor level, NA generated
#> [1] <NA> B <NA> D <NA> F G <NA> I J
#> Levels: A B C D E F G H I J
impute_fixed(vec_fct, "ZZ")
#> Warning in `[<-.factor`(`*tmp*`, is.na(x), value = "ZZ"): invalid factor level, NA generated
#> [1] <NA> B <NA> D <NA> F G <NA> I J
#> Levels: A B C D E F G H I J
However, you can use impute_mode():
impute_mode(vec_fct)
#> [1] I B I D I F G I I J
#> Levels: A B C D E F G H I J
For factors, you can impute a specific value:
impute_factor(vec_fct, value = "ZZ")
#> [1] ZZ B ZZ D ZZ F G ZZ I J
#> Levels: A B C D E F G H I J ZZ
Think of it like impute_fixed(), but a special case for factors.
Now let’s demonstrate how to do this in a data frame. First we create the data
library(dplyr)
dat <- tibble(
num = vec_num,
int = vec_int,
fct = vec_fct
)
dat
#> # A tibble: 10 × 3
#> num int fct
#> <dbl> <int> <fct>
#> 1 NA NA NA
#> 2 0.923 5 B
#> 3 NA 6 NA
#> 4 0.527 5 D
#> 5 NA 5 NA
#> 6 NA NA F
#> 7 1.43 NA G
#> 8 1.38 6 NA
#> 9 -0.786 NA I
#> 10 -0.593 6 J
You can use it inside mutate like so:
dat %>%
mutate(
num = impute_fixed(num, -9999),
int = impute_zero(int),
fct = impute_factor(fct, "out")
)
#> # A tibble: 10 × 3
#> num int fct
#> <dbl> <dbl> <fct>
#> 1 -9999 0 out
#> 2 0.923 5 B
#> 3 -9999 6 out
#> 4 0.527 5 D
#> 5 -9999 5 out
#> 6 -9999 0 F
#> 7 1.43 0 G
#> 8 1.38 6 out
#> 9 -0.786 0 I
#> 10 -0.593 6 J
Or if you want to impute across all applicable variables with a single function, you could use where like so, to focus on numeric data:
dat %>%
mutate(
across(
.cols = where(is.numeric),
.fn = impute_zero
)
)
#> # A tibble: 10 × 3
#> num int fct
#> <dbl> <dbl> <fct>
#> 1 0 0 NA
#> 2 0.923 5 B
#> 3 0 6 NA
#> 4 0.527 5 D
#> 5 0 5 NA
#> 6 0 0 F
#> 7 1.43 0 G
#> 8 1.38 6 NA
#> 9 -0.786 0 I
#> 10 -0.593 6 J
dat %>%
mutate(
across(
.cols = where(is.numeric),
.fn = \(x) impute_fixed(x, -99)
)
)
#> # A tibble: 10 × 3
#> num int fct
#> <dbl> <dbl> <fct>
#> 1 -99 -99 NA
#> 2 0.923 5 B
#> 3 -99 6 NA
#> 4 0.527 5 D
#> 5 -99 5 NA
#> 6 -99 -99 F
#> 7 1.43 -99 G
#> 8 1.38 6 NA
#> 9 -0.786 -99 I
#> 10 -0.593 6 J
Improvements
- Add
digitargument tomiss_var_summaryto help display percentage missing data correctly when there is a very small fraction of missingness - solving #284.
N <- 30000000
df <- tibble(x = rep(NA_real_, N)) %>%
add_row(x = 0)
df %>% miss_var_summary()
#> # A tibble: 1 × 3
#> variable n_miss pct_miss
#> <chr> <int> <num>
#> 1 x 30000000 100.
df %>% miss_var_summary(digits = 6)
#> # A tibble: 1 × 3
#> variable n_miss pct_miss
#> <chr> <int> <num:.6!>
#> 1 x 30000000 99.999997
geom_miss_point()works withshapeargument #290
ggplot(
airquality,
aes(
x = Ozone,
y = Solar.R,
shape = factor(Month)
)
) +
geom_miss_point(size = 4)
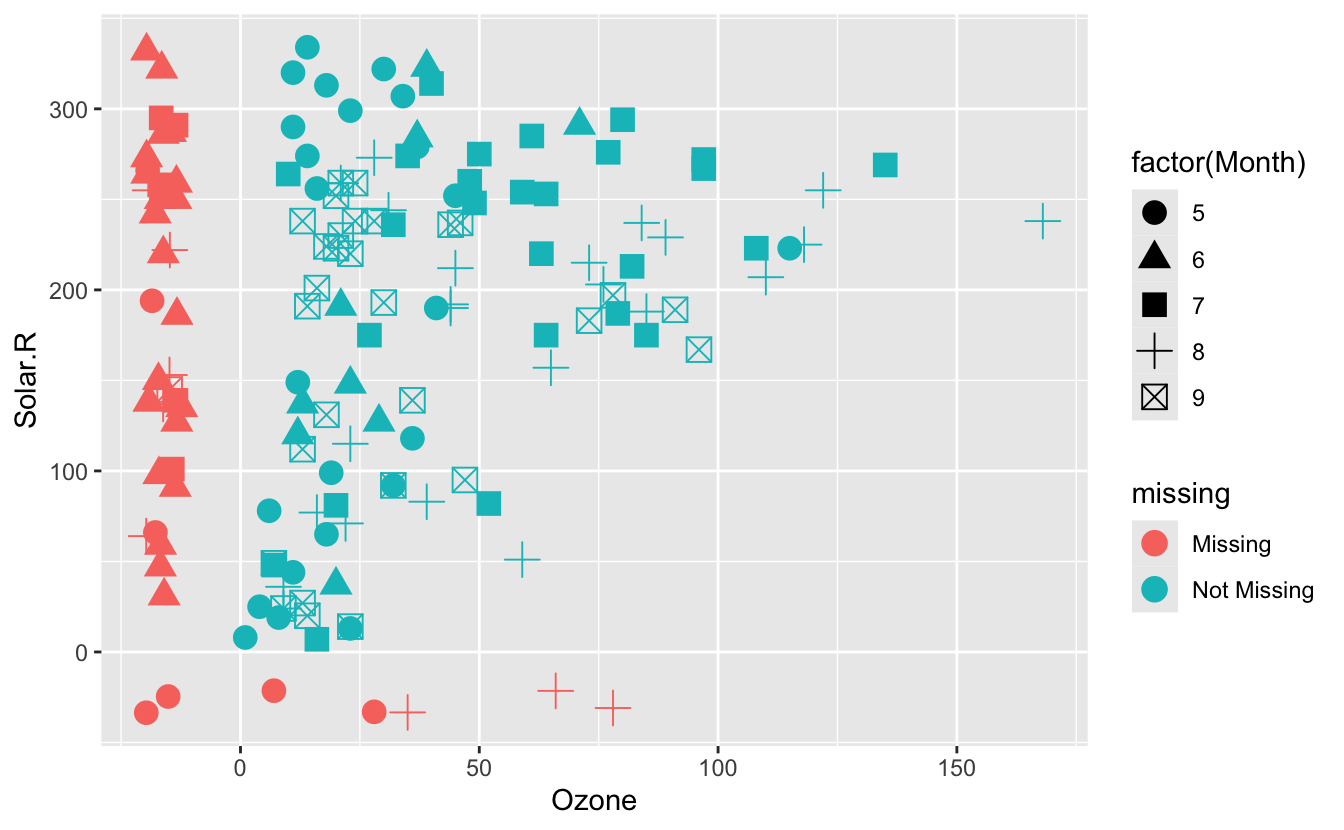
- Implemented
Date,POSIXctandPOSIXltmethods forimpute_below()- #158
dat_date <- tibble(
values = 1:7,
number = c(111, 112, NA, NA, 108, 150, 160),
posixct = as.POSIXct(number, origin = "1970-01-01"),
posixlt = as.POSIXlt(number, origin = "1970-01-01"),
date = as.Date(number)
)
dat_date
#> # A tibble: 7 × 5
#> values number posixct posixlt date
#> <int> <dbl> <dttm> <dttm> <date>
#> 1 1 111 1970-01-01 10:01:51 1970-01-01 10:01:51 1970-04-22
#> 2 2 112 1970-01-01 10:01:52 1970-01-01 10:01:52 1970-04-23
#> 3 3 NA NA NA NA
#> 4 4 NA NA NA NA
#> 5 5 108 1970-01-01 10:01:48 1970-01-01 10:01:48 1970-04-19
#> 6 6 150 1970-01-01 10:02:30 1970-01-01 10:02:30 1970-05-31
#> 7 7 160 1970-01-01 10:02:40 1970-01-01 10:02:40 1970-06-10
date_date_imp <- dat_date %>%
mutate(
number_imp = impute_below(number),
.after = number
) %>%
mutate(
posixct_imp = impute_below(posixct),
.after = posixct
) %>%
mutate(
posixlt_imp = impute_below(posixlt),
.after = posixlt
) %>%
mutate(
date_imp = impute_below(date),
.after = date
)
date_date_imp
#> # A tibble: 7 × 9
#> values number number_imp posixct posixct_imp
#> <int> <dbl> <dbl> <dttm> <dttm>
#> 1 1 111 111 1970-01-01 10:01:51 1970-01-01 10:01:51
#> 2 2 112 112 1970-01-01 10:01:52 1970-01-01 10:01:52
#> 3 3 NA 103. NA 1970-01-01 10:01:43
#> 4 4 NA 102. NA 1970-01-01 10:01:41
#> 5 5 108 108 1970-01-01 10:01:48 1970-01-01 10:01:48
#> 6 6 150 150 1970-01-01 10:02:30 1970-01-01 10:02:30
#> 7 7 160 160 1970-01-01 10:02:40 1970-01-01 10:02:40
#> # ℹ 4 more variables: posixlt <dttm>, posixlt_imp <dttm>, date <date>,
#> # date_imp <date>
Bug fixes
- Fix bug with
all_complete(), which was implemented as!anyNA(x)but should beall(complete.cases(x)). - Correctly implement
any_na()(andany_miss()) andany_complete(). Rework examples to demonstrate workflow for finding complete variables. - Fix bug with
shadow_long()not working when gathering variables of mixed type. Fix involved specifying a value transform, which defaults to character. #314 - Provide
replace_na_with(), a complement toreplace_with_na()- #129
x <- c(1:5, NA, NA, NA)
x
#> [1] 1 2 3 4 5 NA NA NA
replace_na_with(x, 0)
#> [1] 1 2 3 4 5 0 0 0
dat <- tibble(
ones = c(1,1,NA),
twos = c(NA,NA, 2),
threes = c(NA, NA, NA)
)
dat %>%
mutate(
ones = replace_na_with(ones, 0),
twos = replace_na_with(twos, -2),
threes = replace_na_with(threes, -3)
)
#> # A tibble: 3 × 3
#> ones twos threes
#> <dbl> <dbl> <dbl>
#> 1 1 -2 -3
#> 2 1 -2 -3
#> 3 0 2 -3
dat %>%
mutate(
across(
everything(),
\(x) replace_na_with(x, -99)
)
)
#> # A tibble: 3 × 3
#> ones twos threes
#> <dbl> <dbl> <dbl>
#> 1 1 -99 -99
#> 2 1 -99 -99
#> 3 -99 2 -99
- Fix bug with
gg_miss_fct()where it used a deprecated function from forcats - #342
Internal changes
{naniar} now uses cli::cli_abort() and cli::cli_warn() instead of stop() and warning() (#326). Internally in tests we changed to use expect_snapshot() instead of expect_error().
Deprecations
The following functions have been soft deprecated, and will eventually be made defunct in future versions of naniar. shadow_shift() - #193 in favour of impute_below(), and miss_case_cumsum() and miss_var_cumsum() - #257 - in favour of miss_case_summary(data, cumsum = TRUE) and miss_var_summary(data, cumsum = TRUE).
Thanks!
Thanks to everyone for using {naniar}, I’m happy that I’ve got another release out the door and am looking forward to more changes in the future. Especially thanks to everyone who contributed to issues and or pull request for this release, including: @szimmer, @HughParsonage, , @siavash-babaei, @maksymiuks, @jonocarroll, @jzadra, @krlmlr
